pacman::p_load(influence.ME,
lattice, # dotplot
dplyr,
texreg,
stargazer,
tidyr
)Observaciones influyentes
Correspondiente a la sesión del jueves, 5 de junio de 2025
1. Casos influyentes Multinivel
Datos (incluidos en el paquete influence.ME)
- 23 escuelas, 519 individuos
- variables principales:
- Dependiente: rendimiento en matemática
- Independiente L1: homework, tiempo realizando tareas
- Independiente L2: structure, grado de estructura de las clases
data(school23)
dim(school23)
str(school23$homework)
school23 <- within(school23, homework <- unclass(homework)) # pasar a continua
str(school23$homework)Estimación modelo
m23 <- lmer(math ~ homework + structure + (1 | school.ID),data=school23)
summary(m23, cor=FALSE)Linear mixed model fit by REML ['lmerMod']
Formula: math ~ homework + structure + (1 | school.ID)
Data: school23
REML criterion at convergence: 3724.5
Scaled residuals:
Min 1Q Median 3Q Max
-2.6617 -0.7063 -0.0151 0.6665 3.2326
Random effects:
Groups Name Variance Std.Dev.
school.ID (Intercept) 19.54 4.421
Residual 71.31 8.445
Number of obs: 519, groups: school.ID, 23
Fixed effects:
Estimate Std. Error t value
(Intercept) 52.2355 5.3948 9.683
homework 2.3938 0.2771 8.640
structure -2.0950 1.3239 -1.582Plot descriptivo
struct <- school23 %>% select(school.ID, structure, math) %>% group_by(school.ID) %>% summarise_all(mean) %>% as.data.frame()
dotplot(math ~ factor(structure), # lattice
struct,
type=c("p","a"),
xlab="Class structure level",
ylab="Average Math Test Score")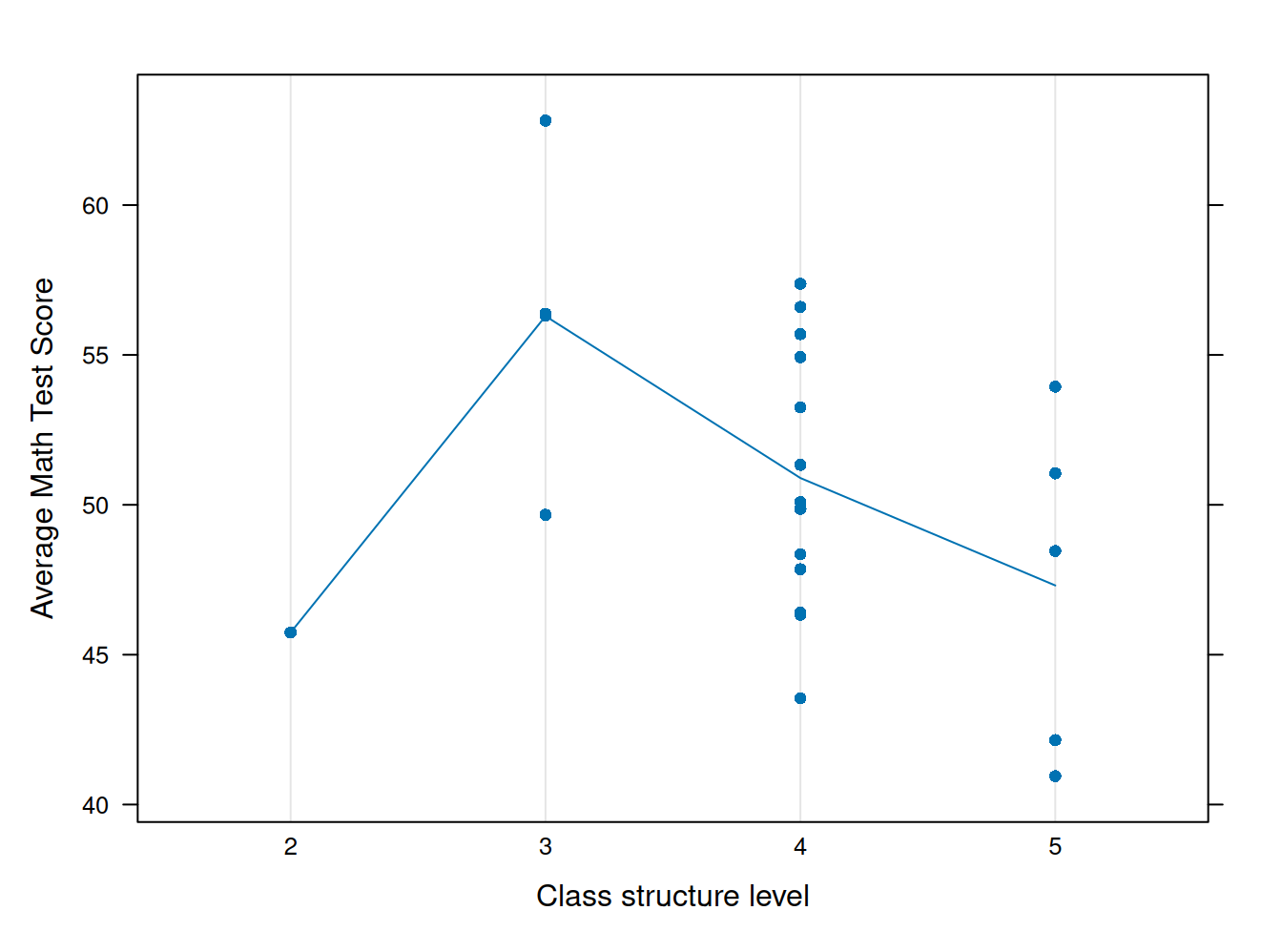
Calculando medidas de influencia basados en función influence de influence.ME
estex.m23 <- influence(m23, "school.ID") Genera objeto estex.m23 para futuras estimaciones de influencia
DFBETAS
dfbetas(estex.m23) (Intercept) homework structure
6053 0.20438272 -0.13353782 -0.168112863
6327 0.11706086 -0.44769047 0.020480558
6467 -0.01647348 0.21089491 0.015320693
7194 0.03155659 -0.44641518 0.036749233
7472 -1.20542747 -0.55839049 1.254798224
7474 -0.22112732 -0.51980380 0.353431371
7801 0.07862924 -0.45872039 -0.012145337
7829 0.58739143 -0.77957141 -0.555013123
7930 -0.09131583 0.65291967 0.019638096
24371 -0.07478227 0.37515821 -0.002611182
24725 -0.14550292 0.60660357 -0.005518191
25456 0.07021527 -0.36410988 -0.005086597
25642 0.02256001 -0.21373926 -0.013416556
26537 0.06514836 0.28956822 -0.095167486
46417 -0.01740458 0.26325092 0.023438196
47583 -0.05615965 0.23266306 0.026355535
54344 0.38857969 0.08338389 -0.474978335
62821 0.66202562 -0.92371257 -0.455327896
68448 -0.30385524 0.41369769 0.222734720
68493 -0.05718403 0.25071141 -0.006237296
72080 -0.04604135 0.32986115 0.008072893
72292 -0.11354833 0.62278357 0.003905476
72991 -0.06669580 0.52021276 0.021627589dfbetas(estex.m23,parameters=c(2,3)) homework structure
6053 -0.13353782 -0.168112863
6327 -0.44769047 0.020480558
6467 0.21089491 0.015320693
7194 -0.44641518 0.036749233
7472 -0.55839049 1.254798224
7474 -0.51980380 0.353431371
7801 -0.45872039 -0.012145337
7829 -0.77957141 -0.555013123
7930 0.65291967 0.019638096
24371 0.37515821 -0.002611182
24725 0.60660357 -0.005518191
25456 -0.36410988 -0.005086597
25642 -0.21373926 -0.013416556
26537 0.28956822 -0.095167486
46417 0.26325092 0.023438196
47583 0.23266306 0.026355535
54344 0.08338389 -0.474978335
62821 -0.92371257 -0.455327896
68448 0.41369769 0.222734720
68493 0.25071141 -0.006237296
72080 0.32986115 0.008072893
72292 0.62278357 0.003905476
72991 0.52021276 0.0216275892/sqrt(23) # cut point[1] 0.4170288dfbeta_estex.m32=as.data.frame(dfbetas(estex.m23))
dfbeta_estex.m32 %>% filter(homework > 0.417 | structure > 0.417 ) (Intercept) homework structure
7472 -1.20542747 -0.5583905 1.254798224
7930 -0.09131583 0.6529197 0.019638096
24725 -0.14550292 0.6066036 -0.005518191
72292 -0.11354833 0.6227836 0.003905476
72991 -0.06669580 0.5202128 0.021627589plot(estex.m23, which="dfbetas", parameters=c(2,3), xlab="DFbetaS", ylab="School ID")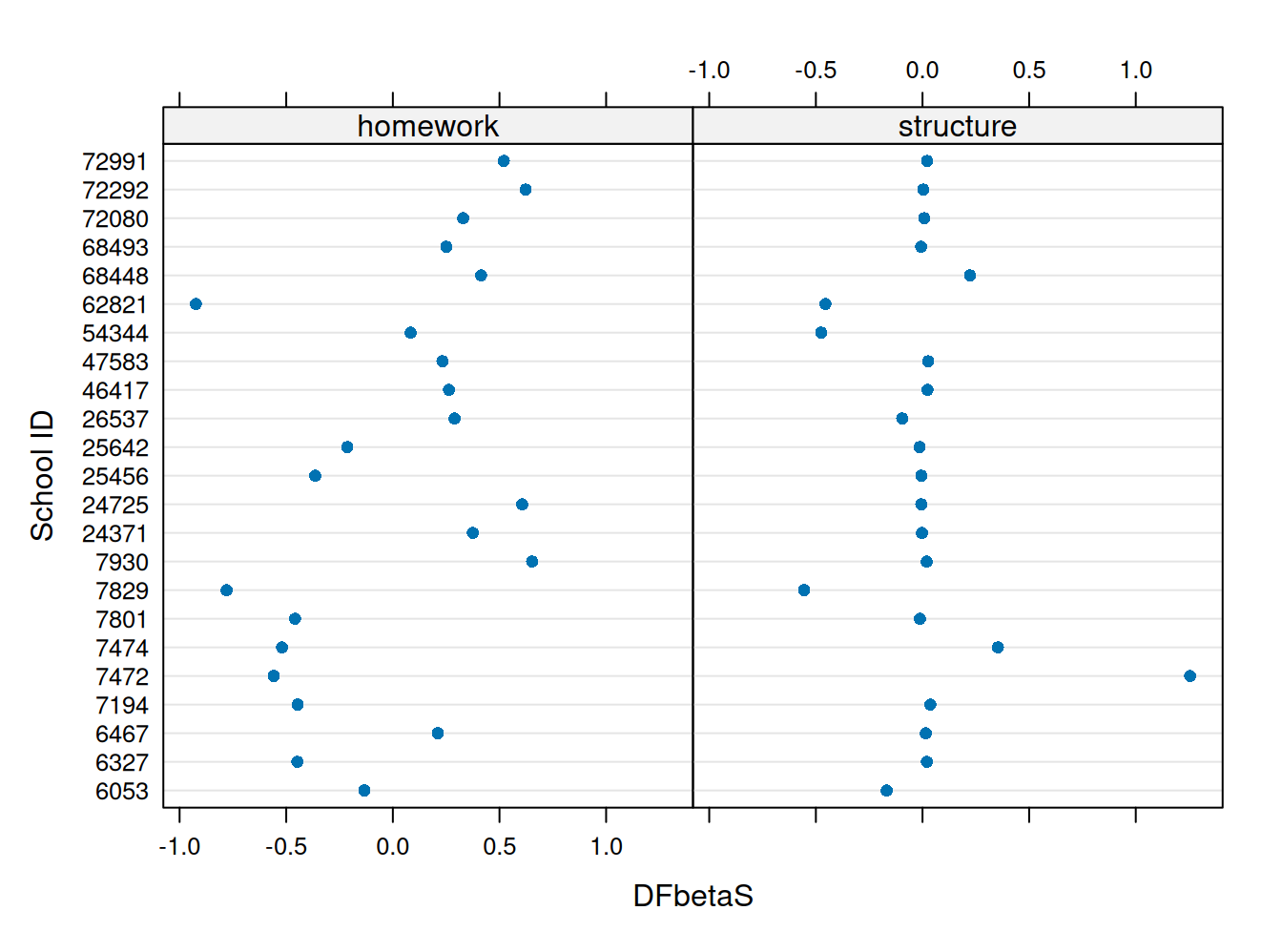
Distancia de Cook
cooks.distance(estex.m23, sort = TRUE)
4/23 # cut pointplot(estex.m23, which="cook",
cutoff=.17, sort=TRUE,
xlab="Cooks Distance",
ylab="School ID")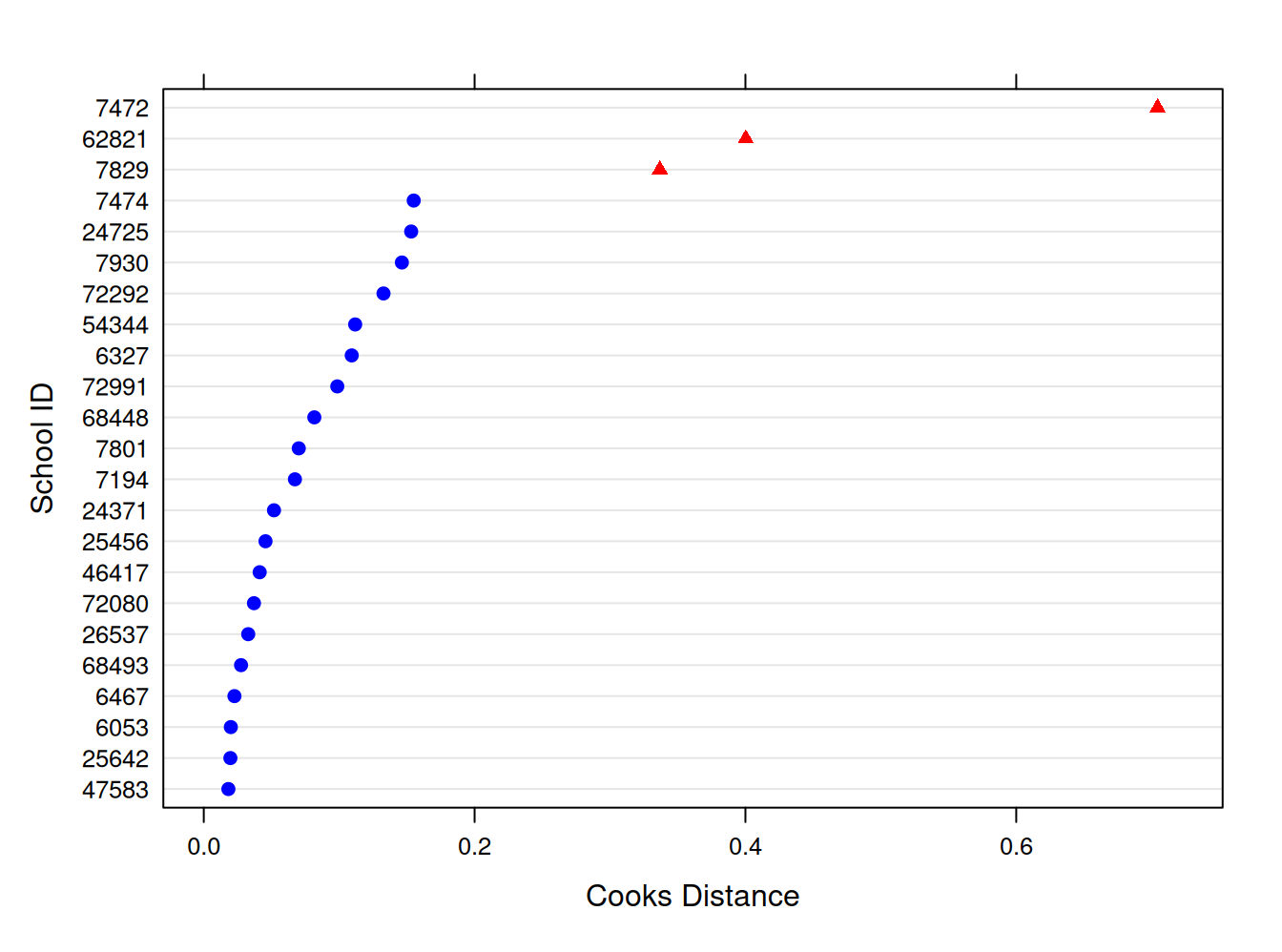
Statistical significance
sigtest(estex.m23, test=-1.96)$structure[1:10,] Altered.Teststat Altered.Sig Changed.Sig
6053 -1.326219 FALSE FALSE
6327 -1.688461 FALSE FALSE
6467 -1.589753 FALSE FALSE
7194 -1.512465 FALSE FALSE
7472 -2.715381 TRUE TRUE
7474 -1.894861 FALSE FALSE
7801 -1.533801 FALSE FALSE
7829 -1.045699 FALSE FALSE
7930 -1.565893 FALSE FALSE
24371 -1.546618 FALSE FALSEPara la variable structure, se muestran las primeras 10 filas, y se establece un criterio de significación de 1.96 para detectar si hay alteraciones en la significación
datab=filter(school23,school.ID!=7472)
m23b <- lmer(math ~ homework + structure + (1 | school.ID),data=datab) sjPlot::tab_model(m23, m23b,
show.ci = FALSE,
dv.labels = c("Modelo original", "Modelo sin influyentes"))| Modelo original | Modelo sin influyentes | |||
|---|---|---|---|---|
| Predictors | Estimates | p | Estimates | p |
| (Intercept) | 52.24 | <0.001 | 59.41 | <0.001 |
| homework | 2.39 | <0.001 | 2.55 | <0.001 |
| structure | -2.10 | 0.114 | -3.89 | 0.007 |
| Random Effects | ||||
| σ2 | 71.31 | 70.67 | ||
| τ00 | 19.54 school.ID | 15.33 school.ID | ||
| ICC | 0.22 | 0.18 | ||
| N | 23 school.ID | 22 school.ID | ||
| Observations | 519 | 496 | ||
| Marginal R2 / Conditional R2 | 0.158 / 0.339 | 0.235 / 0.371 | ||
¿Qué diferencia hay entre el modelo con y sin la observación influyente de nivel 2?